Abstract
Single-cell proteomics can reveal cellular phenotypic heterogeneity and cell-specific functional networks underlying biological processes. Here, we present a streamlined workflow combining microfluidic chips for all-in-one proteomic sample preparation and data-independent acquisition (DIA) mass spectrometry (MS) for proteomic analysis down to the single-cell level. The proteomics chips enable multiplexed and automated cell isolation/counting/imaging and sample processing in a single device. Combining chip-based sample handling with DIA-MS using project-specific mass spectral libraries, we profile on average ~1,500 protein groups across 20 single mammalian cells. Applying the chip-DIA workflow to profile the proteomes of adherent and non-adherent malignant cells, we cover a dynamic range of 5 orders of magnitude with good reproducibility and <16% missing values between runs. Taken together, the chip-DIA workflow offers all-in-one cell characterization, analytical sensitivity and robustness, and the option to add additional functionalities in the future, thus providing a basis for advanced single-cell proteomics applications.
Introduction
Rapidly developing single-cell omics-based molecular measurements have revolutionized modern biological research1,2. As proteins are functional workhorses of the cell, proteomic profiling provides a direct snapshot of the dynamic biological network to complement the genomics and transcriptomics architecture3. However, the sensitivity of proteomic profiling is limited due to the wide dynamic range of proteome constituents and the lack of a viable protein amplification strategy4. Targeted protein analyses have enabled sensitivity down to the single-cell level, but their multiplexity is often limited and depends on antibody availability and quality5,6,7,8. Mass spectrometry (MS)-based proteomic approaches offer label-free analysis with high specificity and deep proteomic coverage, which has been shown in several studies to reach single-cell sensitivity8,9,10,11,12,13,14. However, multistep processing in most traditional MS workflows often results in significant sample loss, linking trade-offs between high proteome coverage and accessible sample size8,15.
Microproteomics workflows aimed at handling minute samples have been widely developed to expand MS-based proteomic analysis toward limited input samples (<1000 cells)16. For example, filter-aided sample preparation (FASP), inStageTip (iST), integrated proteome analysis device (iPAD), and single-pot solid-phase-enhanced sample preparation (SP3) reported protocols that combine cell lysis, protein digestion, and/or detergent removal to improve proteome identification at the level of a few hundred cells17,18,19,20. Alternatively, sample preparations on nanoliter droplets have been developed to enhance proteome profiling sensitivity, including oil-air droplet (OAD) and digital microfluidic (DMF-SP3) chips; these methods effectively reduced adsorptive loss and identified 1063 and 2500 proteins from 100 and 500 cells, respectively21,22. Similarly, the nanodroplet processing platform (nanoPOTS) achieved proteome coverage of over 3000 proteins from 10 to 100 cells by incorporating the match-between-runs (MBR) feature23. Its recent extension with ultralow-flow nanoLC and high-field asymmetric ion mobility spectrometry (FAIMS) coupled to the Orbitrap Eclipse instrument reported sensitive profiling of 1475 protein groups with MBR from a single cell13. Incorporation of isotopic labeling to extend the multiplexity ability was demonstrated at the level of 1000 proteins in primary cells12,24. With these advances, nonetheless, a fully automated workflow, starting from multiplexed cell capturing and imaging, cell lysis, and protein digestion to peptide desalting, all integrated within a single device to realize proteomic analysis for low-input samples has not yet been established, despite the prospect to substantially minimize sample loss and achieve high reproducibility and sensitivity.
Microfluidic devices based on multilayer soft lithography use custom chip integration and hydraulic actuations to achieve precise μL-to-nL fluid manipulation and are ideal platforms to execute a complex protocol25,26,27. However, microfluidics has not been explored for streamlined proteomics workflows primarily due to challenges associated with reagent compatibility for one-pot protocols, concerns of mixing in confined space, and overall system integration. To study the cellular or phenotypic status, sufficient proteome coverage is critical and may require higher cell numbers due to relatively limited proteome coverages at the single-cell level. Therefore, in this study, chips with different cell capacities were constructed to facilitate experiments with optimal profiling depth for different cell inputs. Specifically, an integrated proteomics chip (iProChip, 1–100 cells) and its extended version for single-cell capacity (SciProChip) were designed and coupled with data-independent acquisition (DIA) MS as streamlined nanoproteomics (nanogram of cells) pipelines. These chips are designed as automated stations for the entire proteomic workflow, offering built-in features including quantifiable cell capture and imaging, complete cell lysis, protein digestion, and peptide desalting. In situ cell counting allows quantification of the number of captured cells. Thus, the cell number can be variable to achieve proteome coverage of interest. Following chip processing, we showed that DIA MS, which detects all precursors and fragments in the entire m/z range within isolation windows, enabled all retrospective peptide mapping against spectral libraries and offered 2.3-fold superior coverage than conventional data-dependent acquisition (DDA) mode28. Importantly, the SciProChip-DIA workflow characterized 1500 ± 131 protein groups from individual single cells with 1% false discovery rate (FDR). Furthermore, the analytical performance and versatility of iProChip-DIA were demonstrated using both human adenocarcinoma cells (PC-9) and chronic B cell leukemia cells (MEC-1), whose size differences were readily quantified using the built-in cell imaging feature. The results revealed a performance of 5 orders of proteome coverage, >100-fold quantification range, good reproducibility (Pearson correlation of 0.88–0.98) and low between-run missing values (<16%). The presented workflow illustrates a unique implementation of microfluidic devices with all-in-one functionality to achieve automated and streamlined proteomic preparation, which offers high sensitivity and reproducibility for limited input samples, including a single cell.
Results
Design and characterization of the iProChip and streamlined microproteomics workflow
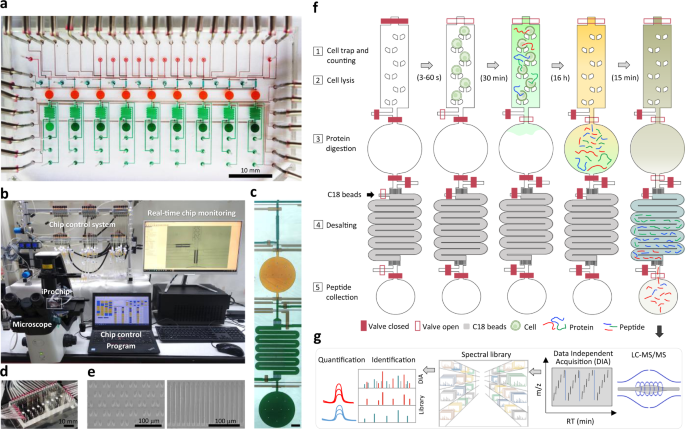
To provide a streamlined microproteomic pipeline for mass-limited samples, we designed a microfluidic device as an integrated proteomics chip (iProChip) to offer all-in-one functionality from cell input to complete proteomic sample processing. The iProChip has a two-layer, push-up geometry and allows accurate fluid manipulation via 34 valves controlled by a custom program, thereby offering an automated protocol for precise and systematic control (Fig. 1a–f and Supplementary Fig. 1a)29. The chip is composed of 9 units to enable multiplexed proteomic experiments running in parallel. Each unit contains a cell capture, imaging and lysis chamber, a protein reduction, alkylation and digestion vessel, and a peptide desalting column (Fig. 1c and Supplementary Fig. 1b). All units share 9 inlets and 2 outlets, allowing programmed delivery of reagents and simultaneous sample processing to increase assay throughput. The cell trap is made up of arrays of 10, 50 and 100 wedge-shaped twin pillars spaced by 5 μm for rapid size-based cell capture in 5, 8.5, and 11 nL chambers, respectively (Fig. 1e)30. A circular chamber with a radius of 1 mm and a height of 100 μm (312 nL) was fabricated to accommodate the entire proteomic workflow, including cell lysis, protein reduction, alkylation and digestion in a single step (Fig. 1f and Supplementary Fig. 2). Note that the calculated surface-to-volume ratio for iProChip is larger than that of existing single-cell devices, such as nanoPOTS, yet it still exhibits a substantially reduced surface area by >90% in comparison to the microscale vial-based workflow (Supplementary Table 1)11. For peptide desalting, a 2.5 cm-long column with a cross-section of 200 μm × 25 μm was fabricated by packing reversed-phase C18 beads into the microchannel prepatterned with 5 μm filters to perform on-chip clean-up of digested peptides (Fig. 1e, f and Supplementary Movie 1). To increase proteome coverage, we applied a deep single-shot profiling strategy that integrates direct- and library-based DIA analysis using an Orbitrap mass spectrometer. We developed a spectral library resource complementarily established by hybrid DDA-DIA datasets using either cancer cell lines or immune cells consisting of different proteome compositions, which can serve as a digital map to theoretically recover all peptides in the m/z and retention time domains of DIA data (Fig. 1g). Specifically, spectral libraries constructed from cell lines with different cell numbers were tested and optimized to maximize the number of identified and quantified proteins.
a A bright-field image of the integrated proteomics chip (iProChip), where cell capture chambers (cyan), reaction vessels (orange), on-chip SPE columns (green), sample collection ports (dark green), and control layers (brown) are shown. b The entire system set-up for iProChip operation. c A close-up view of a single operation unit. Scale bar: 300 μm. d A ready-to-use iProChip mounted on the microscope. e SEM images of cell capturing pillars (left) and C18 filters in the SPE column (right). These images are representative of two chips that were observed using SEM. f Operational procedures of iProChip for streamlined sample preparation, including (1) cell trapping, imaging, and counting, (2) cell lysis, (3) protein digestion, (4) desalting, and (5) peptide collection. g Proteomic analysis using data-independent acquisition-based liquid chromatography–tandem mass spectrometry (LC-MS/MS) and spectral library search.
In the first step of the streamlined workflow, the cell trapping efficiency was determined using non-small-cell lung cancer (NSCLC) PC-9 cells (“Methods”). Using optimal cell density (500 cells/μL), desired numbers of cells (1–100) for each unit can be trapped in 10–60 s. The average percentage of cells captured from traps containing a single cell were 100, 92 ± 3, and 89 ± 8% for chambers with 10, 50, and 100 traps, respectively. The targeted capture efficiency for all units reached ~100% after counting traps containing 1 (~90%) and 2 or 3 cells (~10%), establishing it as an absolute quantifiable module to perform simple and fast size-based cell isolation (Fig. 2a, b, “Methods,” and Supplementary Movie 2). Compared to external stand-alone cell sorters, such a built-in module offers simple, rapid, and efficient cell isolation. Additionally, we also showed that by using a lower cell density (25 cells/μL) operated at 3 psi, such cell chambers allow precise capture of lower numbers of cells at the level of 1 and 5 cells (Supplementary Fig. 3). The cell trapping capability of iProChip was also evaluated to characterize the cell usage efficiency (defined as numbers of trapped cells/numbers of total injected cells) and minimum numbers of cells needed for iProChip operation (“Methods”). Using a total of either 5 or 10 μL cell solution (25 cells/μL), the results showed that cell usage efficiency ranged from ~4 to 44% for capturing 1–100 cells (Supplementary Table 2 and Supplementary Fig. 4). Next, we sought to characterize whether reagents can mix efficiently in the closed vessel during cell lysis and protein digestion. Three mixing approaches, including vortexing, shaking (by a plate shaker), and passive diffusion, were tested (Supplementary Fig. 5). Using imaging analysis, the relative mixing index (RMI) was calculated to assess the mixing performance (“Methods”)31. The results showed that it took 11, 16, and 30 min for vortexing, shaking, and diffusion-mixing to reach 75% RMI, indicating that all three mixing strategies were sufficient to accommodate reactions within minutes to hours of reaction kinetics, which fit the timescale of conducting the proteomics workflow (Fig. 2c, d and Supplementary Fig. 5). Although vortex mixing was found to provide faster mixing, mixing by shaking was used in subsequent experiments due to its flexibility in handling and sufficient reaction timescale.