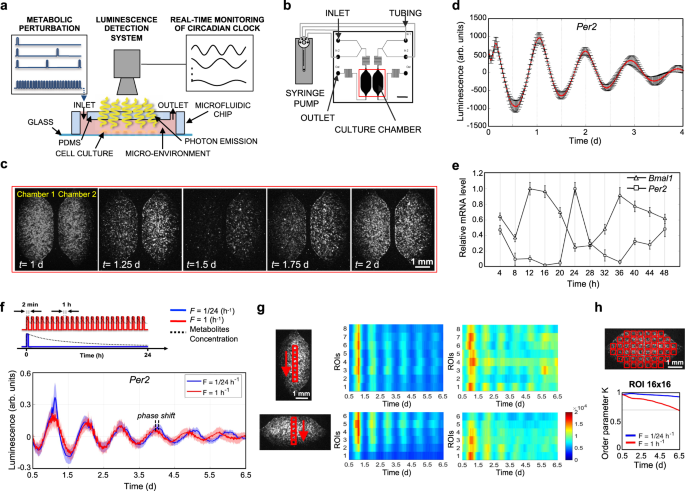
We designed and developed a microfluidic platform (Supplementary Fig. S7) to perform frequency-encoded metabolic perturbations, while recording circadian gene expression by luminescence detectors (Fig. 1a). This microfluidic setup was connected with an external liquid handling system for accurate control of medium delivery (Fig. 1b). The recording of luminescence signals from the two 1.75 μL microfluidic cell culture chambers was performed under a temperature-controlled microscope providing a spatial resolution of 20 μm.
a Schematic representation of experimental setup for the acquisition of bioluminescence signal from cell integrated microfluidic chip; microfluidic setup is coupled with a microscope and liquid handling system. b Schematic representation of the liquid handling system connected to the microfluidic device, composed of two independent culture chambers (two inlets and one outlet for each). Scale bar 2.5 mm. c Time-lapse of bioluminescence images of a representative experiment, captured every 30 min, of mouse Per2::Luc fibroblasts, showing circadian rhythms of luminescence for 2 days. Scale bar 1 mm. d Baseline-subtracted Per2::Luc bioluminescence patterns acquired for 4 days from a microfluidic chamber after dexamethasone shock. Data are represented as mean ± s.d., N = 9 for each condition. e Temporal profiles of clock gene expressions, BMAL1 and PER2, in human fibroblasts integrated in the microfluidic platform, after synchronization (t = 0) with dexamethasone. mRNA level of clock genes was measured by qPCR and normalized to GADPH expression. Data are represented as mean ± s.d., N = 3 for each group. f Top, Schematic representation of the two different protocols of medium delivery imposed by automatic medium change. Bottom, Per2::Luc 6-day bioluminescence signal intensity in the 16-by-16 pixel ROIs, shown in the scheme in h (top), under the two medium change conditions, after baseline subtraction using LumiCycle Analysis program (Actimetrics). Solid line: mean signal intensity, patch: area delimited by mean ± s.d. of the 48 ROIs indicated in h. g Heatmaps of mean Per2::Luc 6-day bioluminescence signal intensity in the 16-by-16 pixel ROIs, shown in the schemes on the left, under the two medium change conditions shown in f. Colorbar indicates the bioluminescence signal intensity after image pre-processing in arbitrary units and applies to all four heatmaps. Scale bar 1 mm. h Kuramoto order parameter K, calculated for the time intervals between time 0.5 d and the time shown on the x axis, indicating the level of synchrony between the 16-by-16 pixel ROIs, shown in the scheme on top (K = 1 means complete synchrony). The horizontal gray line highlights a value of 0.7. Source data are available as a Source Data file. Scale bar 1 mm.
As proof of concept of circadian synchronization of mammalian cell cultures in a microfluidic environment, Fig. 1c and Supplementary Movie 1 show a time-lapse of luminescence signal from mouse Per2::Luc fibroblasts. The microfluidic experimental setup permits recording of highly reproducible, robust, and spatially resolved oscillatory behavior of the cell population after dexamethasone shock (Fig. 1d). To further verify the feasibility of microfluidic circadian investigation, we performed a qPCR analysis of Per2 and Bmal1 gene expression in Human Foreskin Fibroblasts (HFF) for 48 h after dexamethasone shock (Fig. 1e), a typical protocol for in vitro circadian entrainment46. The expression of the two genes was rhythmic and in anti-phase of 12 h, according to the mechanisms of transcription/translation feedback loops that regulate the circadian clock at the molecular level. These results showed the feasibility of circadian studies using microfluidic technology.
The capability to manipulate and control the microfluidic cellular microenvironment by external liquid handling offers unlimited strategies of how cell culture can be perfused, from low to high frequency of periodic perfusion47. However, how the perfusion frequency affects the circadian clock needs to be investigated, in consideration of a balance between nutrient delivery and waste product removal or a spatial heterogeneity that perfusion strategies can avoid. Here, we implemented two protocols of frequency of medium delivery, F = 1/24 h−1 and F = 1 h−1 (Fig. 1f, top), that were simultaneously imaged in two independent microfluidic cell culture chambers (Fig. 1c). A flow rate of 3 μL/min was used to minimize the shear stress on the cell culture47. The temporal patterns of Per2::Luc mouse fibroblasts exhibited robust circadian expression for 6.5 days in both conditions, as shown in Fig. 1f. Interestingly, ultradian medium changes led to a shorter period of 22.6 ± 0.8 h, compared to 23.7 ± 0.3 h obtained with F = 1/24 h−1 (Supplementary Fig. S2a). We observed a consistent increase of the phase shift, measured as a time difference between two corresponding peaks in the two protocols (F = 1/24 h−1; F = 1 h−1), from 0.1 ± 1.8 h at day 1 to 6.3 ± 5.7 h at day 6 (Supplementary Fig. S2b).
To investigate the phase without any assumption of constant-period oscillatory behavior in either medium change regime, we analyzed the instantaneous phase using Hilbert transform43 (Supplementary Fig. S2d, e). The parallel trends in both plots in Supplementary Fig. S2d demonstrate the constant-period hypothesis as reasonable, however, higher variability in the phase of different ROIs in F = 1 h−1 condition is showed in Supplementary Fig. S2e.
In order to ensure that circadian behavior was homogeneous throughout the cell culture surface and was independent from the specific region of the microfluidic cell culture chamber (typically, upstream/downstream or central/lateral position), we analyzed the images of the two culture chambers by discretizing the vertical and horizontal directions into adjacent ROIs of 16 × 16 pixels (Fig. 1g).
We found that the protocol with F = 1/24 h−1 ensured a more homogeneous distribution of the luminescence signal in horizontal and vertical directions, proving a well-defined circadian behavior. Conversely, the protocol with F = 1 h−1 was able to keep a good synchronization until day 2, after which the circadian gene expression became overall desynchronized.
In order to quantitatively analyze the synchronization over the whole microfluidic surface, we calculated the Kuramoto order parameter42,44 for 16 × 16 pixel ROIs. The results in Fig. 1h confirmed that F = 1/24 h−1 provided a higher degree of spatial synchronization compared to F = 1 h−1. This result is independent from the ROI size as reported in Supplementary Fig. S2f, showing results for 8 × 8 and 32 × 32 pixel ROI.
In general, we can assume that the higher frequency of medium delivery (F = 1 h−1) led to a constant concentration of exogenous factors during an entire 24 h cycle (quasi-steady-state high-glucose condition). Whereas F = 1/24 h−1 induced dynamic changes of exogenous factors because of cellular uptake during 24 h cycle, which closely mimics typical oscillatory day/night behavior. Accordingly, we simulated this scenario with a simplified model of a representative metabolite concentration (Fig. 1f top).